Window Functions CTEs Lab
window functions, CTEs, trees database, data quality
dataset
We’ll work on the normalized version of the trees db
-
download the sql backup from https://skatai.com/assets/db/treesdb_v03_normalized.sql
-
restore the database
from the terminal
psql -d postgres -f <path to the sql backup>/treesdb_v03_normalized.sql
or within a PSQL session
\i <path to the sql backup>/treesdb_v03_normalized.sql
This will create a new database called treesdb_v03_normalized
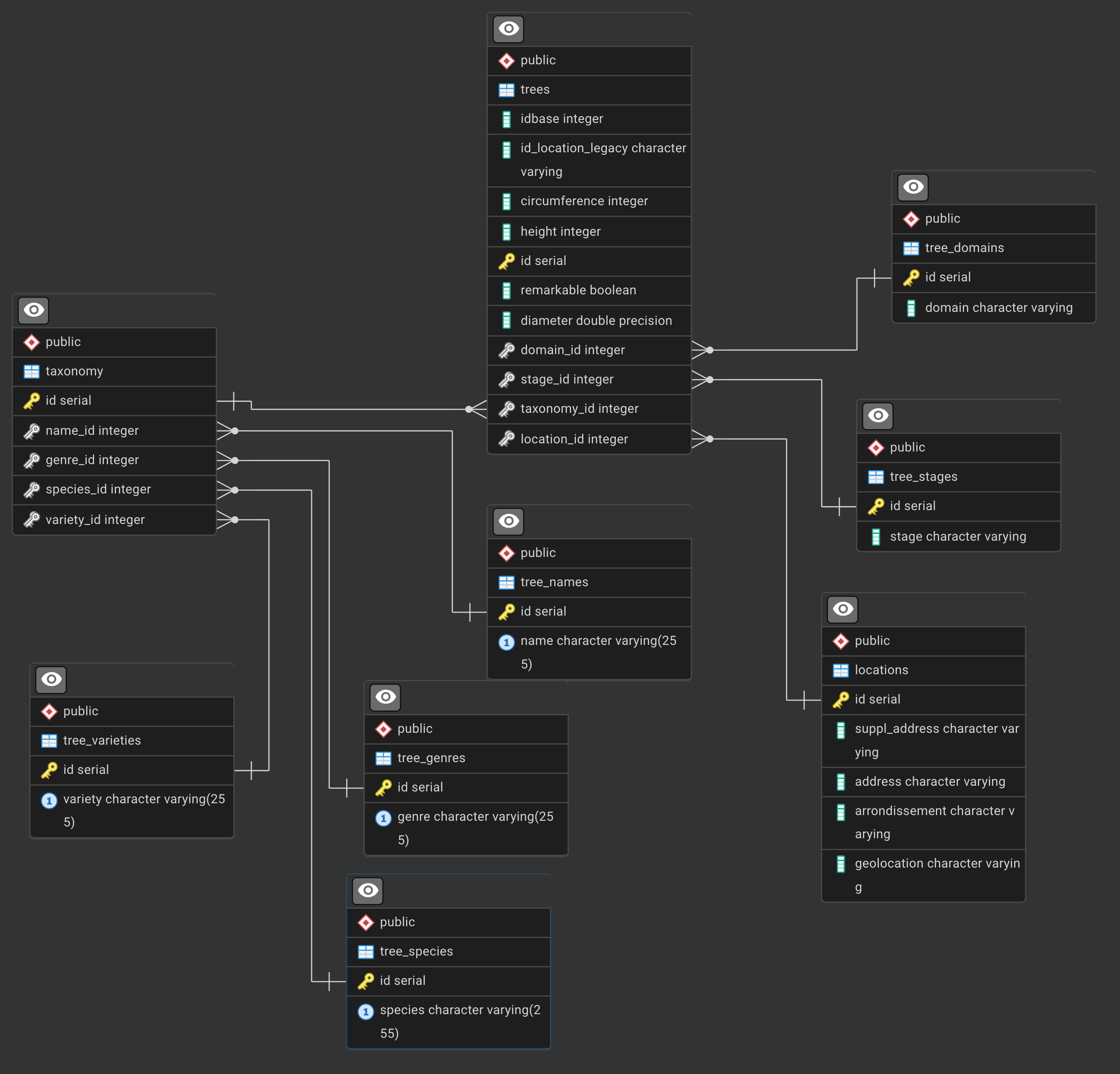
Explore
Check out the tables, make a few queries to get a sense of the data.
The ERD is
PostgreSQL Window Functions Exercises: Paris Trees Database
This comprehensive series of exercises will help you master window functions in PostgreSQL using a real-world dataset of trees in Paris.
important: all results are obtained after ordering by random() and limiting to a few rows. all your queries should also be ordered by random() and limited to a few rows.
Exercise 1: Basic Window Functions - Tree Count by Arrondissement
Goal: For each tree, show its arrondissement and the total count of trees in that arrondissement.
Concepts: Basic COUNT() window function with OVER(PARTITION BY)
Step-by-step guide:
- Start with the main
treestable - Join with
locationsto get arrondissement information - Use
COUNT(*) OVER (PARTITION BY arrondissement)to count trees per arrondissement - Select the tree id, arrondissement, and the count
Hints:
- Window functions don’t reduce rows like
GROUP BYdoes - Each tree row will show the total count for its arrondissement
- The
PARTITION BYclause divides the data into groups for the window function
For comparison - GROUP BY approach:
results
id | arrondissement | trees_in_arrondissement
--------+-------------------+-------------------------
108631 | PARIS 16E ARRDT | 17128
208989 | PARIS 1ER ARRDT | 1645
18713 | PARIS 13E ARRDT | 17462
159547 | PARIS 19E ARRDT | 15235
Solution:
SELECT
t.id,
l.arrondissement,
COUNT(*) OVER (PARTITION BY l.arrondissement) as trees_in_arrondissement
FROM trees t
JOIN locations l ON t.location_id = l.id
WHERE l.arrondissement IS NOT NULL
ORDER BY trees_in_arrondissement DESC, l.arrondissement;
SELECT
l.arrondissement,
COUNT(*) as tree_count
FROM trees t
JOIN locations l ON t.location_id = l.id
WHERE l.arrondissement IS NOT NULL
GROUP BY l.arrondissement
ORDER BY tree_count DESC;
Exercise 2: Finding the Tallest Tree per Arrondissement
Goal: For each tree, show how its height compares to the maximum height in its arrondissement.
Concepts: MAX() window function, comparing values to aggregates
Step-by-step guide:
- Join
treesandlocationstables - Use
MAX(height) OVER (PARTITION BY arrondissement)to find the tallest tree per area - Calculate the difference between each tree’s height and the maximum
- Filter to only show trees with valid heights
Hints:
- You can use multiple columns in your SELECT with window functions
- Consider what
height - MAX(height) OVER (...)tells you - Trees with a difference of 0 are the tallest in their arrondissement
to get a better view of the results, you can limit the number of rows returned and order by random()
ORDER BY random() <other order by> limit 20;
Solution:
SELECT
t.id,
l.arrondissement,
t.height,
MAX(t.height) OVER (PARTITION BY l.arrondissement) as max_height_in_area,
t.height - MAX(t.height) OVER (PARTITION BY l.arrondissement) as diff_from_max
FROM trees t
JOIN locations l ON t.location_id = l.id
WHERE t.height IS NOT NULL
AND l.arrondissement IS NOT NULL
ORDER BY random() , l.arrondissement, diff_from_max DESC limit 20;
results
id | arrondissement | height | max_height_in_area | diff_from_max
--------+-------------------+--------+--------------------+---------------
5422 | PARIS 11E ARRDT | 35 | 35 | 0
133998 | PARIS 20E ARRDT | 15 | 30 | -15
61004 | BOIS DE VINCENNES | 10 | 120 | -110
86518 | PARIS 20E ARRDT | 12 | 30 | -18
Exercise 3: Data Quality - Missing Heights by Genre
Here’s the corrected query for Exercise 3:
Exercise 3: Data Quality - Missing Heights by Genre
Goal: Identify genres with missing height data by showing the count and percentage of trees with missing (0) heights.
Concepts: COUNT() with conditions, calculating percentages with window functions
Step-by-step guide:
- Join trees with taxonomy and tree_genres tables
- Use
COUNT(*) OVER (PARTITION BY genre)for total trees per genre - Use
COUNT(CASE WHEN height > 0 THEN 1 END) OVER (PARTITION BY genre)for trees with valid heights - Calculate the difference to find missing values (height = 0)
- Compute percentage of missing data
Hints:
- In this dataset, missing heights are stored as 0, not NULL
COUNT(CASE WHEN height > 0 THEN 1 END)only counts trees with actual height valuesCOUNT(*)counts all rows- The difference gives you the count of missing/zero heights
- Use
DISTINCTin the outer query to avoid repeated calculations
Solution:
SELECT DISTINCT
tg.genre,
COUNT(*) OVER (PARTITION BY tg.genre) as total_trees,
COUNT(CASE WHEN t.height > 0 THEN 1 END) OVER (PARTITION BY tg.genre) as trees_with_height,
COUNT(CASE WHEN t.height = 0 THEN 1 END) OVER (PARTITION BY tg.genre) as missing_height,
ROUND(100.0 * COUNT(CASE WHEN t.height = 0 THEN 1 END) OVER (PARTITION BY tg.genre) /
COUNT(*) OVER (PARTITION BY tg.genre), 2) as pct_missing
FROM trees t
JOIN taxonomy tx ON t.taxonomy_id = tx.id
JOIN tree_genres tg ON tx.genre_id = tg.id
ORDER BY pct_missing DESC;
Alternative approach (more explicit):
SELECT DISTINCT
tg.genre,
COUNT(*) OVER (PARTITION BY tg.genre) as total_trees,
COUNT(*) OVER (PARTITION BY tg.genre) - COUNT(CASE WHEN t.height = 0 THEN 1 END) OVER (PARTITION BY tg.genre) as trees_with_height,
COUNT(CASE WHEN t.height = 0 THEN 1 END) OVER (PARTITION BY tg.genre) as missing_height,
ROUND(100.0 * COUNT(CASE WHEN t.height = 0 THEN 1 END) OVER (PARTITION BY tg.genre) /
COUNT(*) OVER (PARTITION BY tg.genre), 2) as pct_missing
FROM trees t
JOIN taxonomy tx ON t.taxonomy_id = tx.id
JOIN tree_genres tg ON tx.genre_id = tg.id
ORDER BY pct_missing DESC;
results
genre | total_trees | trees_with_height | missing_height | pct_missing
-------------------+-------------+-------------------+----------------+-------------
Rhododendron | 8 | 0 | 8 | 100.00
Podocarpus | 2 | 0 | 2 | 100.00
Tsuga | 1 | 0 | 1 | 100.00
Cephalotaxus | 1 | 0 | 1 | 100.00
Juniperus | 5 | 0 | 5 | 100.00
Cercidiphyllum | 8 | 1 | 7 | 87.50
Note: a SQL query with
GROUP BYwould be simpler,
For instance
SELECT
tg.genre,
COUNT(*) as total_trees,
COUNT(CASE WHEN t.height > 0 THEN 1 END) as trees_with_height,
COUNT(CASE WHEN t.height = 0 THEN 1 END) as missing_height,
ROUND(100.0 * COUNT(CASE WHEN t.height = 0 THEN 1 END) / COUNT(*), 2) as pct_missing
FROM trees t
JOIN taxonomy tx ON t.taxonomy_id = tx.id
JOIN tree_genres tg ON tx.genre_id = tg.id
GROUP BY tg.genre
ORDER BY pct_missing DESC;
Exercise 4: ROW_NUMBER - Ranking Trees by Height within Each Domain
Goal: Assign a sequential ranking to trees by height within each domain.
Concepts: ROW_NUMBER(), ordering within partitions
Step-by-step guide:
- Join trees with tree_domains
- Use
ROW_NUMBER() OVER (PARTITION BY domain ORDER BY height DESC) - Filter for trees that have height values
- Show only the top 5 trees per domain
Hints:
ROW_NUMBER()always gives unique sequential numbers (1, 2, 3, …)- Use a subquery to filter by row_number (you can’t use window functions in WHERE directly)
ORDER BY height DESCputs tallest trees first
Query Skeleton:
SELECT * FROM (
SELECT
...,
ROW_NUMBER() OVER (...) as rn
FROM ...
WHERE ...
) subquery
WHERE rn <= 5;
Solution:
SELECT * FROM (
SELECT
t.id,
td.domain,
t.height,
ROW_NUMBER() OVER (PARTITION BY td.domain ORDER BY t.height DESC) as height_rank
FROM trees t
JOIN tree_domains td ON t.domain_id = td.id
WHERE t.height IS NOT NULL
AND td.domain IS NOT NULL
) ranked
WHERE height_rank <= 5
ORDER BY domain, height_rank;
results
id | domain | height | height_rank
--------+--------------+--------+-------------
172122 | Alignement | 2524 | 1
152402 | Alignement | 2019 | 2
166842 | Alignement | 720 | 3
168045 | Alignement | 225 | 4
93247 | Alignement | 200 | 5
88686 | CIMETIERE | 119 | 1
91418 | CIMETIERE | 116 | 2
91576 | CIMETIERE | 35 | 3
Exercise 5: RANK vs DENSE_RANK - Handling Ties in Circumference
Goal: Compare RANK() and DENSE_RANK() behavior when trees have the same circumference within each arrondissement.
Concepts: RANK(), DENSE_RANK(), understanding ties
Step-by-step guide:
- Join trees with locations
- Apply both
RANK()andDENSE_RANK()with the same ordering - Order by circumference descending within each arrondissement
- Show the difference between the two ranking methods
Hints:
RANK()leaves gaps after ties: 1, 2, 2, 4, 5DENSE_RANK()has no gaps: 1, 2, 2, 3, 4- Look for trees with the same circumference to see the difference
- Limit results to make patterns visible
Solution:
SELECT
t.id,
l.arrondissement,
t.circumference,
RANK() OVER (PARTITION BY l.arrondissement ORDER BY t.circumference DESC) as rank_with_gaps,
DENSE_RANK() OVER (PARTITION BY l.arrondissement ORDER BY t.circumference DESC) as dense_rank_no_gaps,
RANK() OVER (PARTITION BY l.arrondissement ORDER BY t.circumference DESC) -
DENSE_RANK() OVER (PARTITION BY l.arrondissement ORDER BY t.circumference DESC) as gap_size
FROM trees t
JOIN locations l ON t.location_id = l.id
WHERE t.circumference IS NOT NULL
AND l.arrondissement IS NOT NULL
ORDER BY l.arrondissement, t.circumference DESC
LIMIT 100;
Exercise 6: Detecting Outliers - Heights Beyond 2 Standard Deviations
Goal: Identify unusually tall or short trees by comparing them to statistical measures within their species.
Concepts: AVG(), STDDEV() window functions, statistical outlier detection
Step-by-step guide:
- Join trees, taxonomy, and tree_species tables
- Calculate
AVG(height)andSTDDEV(height)per species using window functions - Compute z-score:
(height - avg) / stddev - Filter for trees where absolute z-score > 2
- Use a subquery since you need to filter on calculated window function results
Hints:
- A z-score shows how many standard deviations a value is from the mean
- Z-score > 2 or < -2 typically indicates an outlier
ABS()function gets absolute value- Only include species with enough trees for meaningful statistics
Query Skeleton:
SELECT * FROM (
SELECT
...,
AVG(...) OVER (PARTITION BY ...) as avg_height,
STDDEV(...) OVER (PARTITION BY ...) as stddev_height,
(height - AVG(...) OVER (...)) / STDDEV(...) OVER (...) as z_score
FROM ...
WHERE ...
) stats
WHERE ABS(z_score) > 2
AND stddev_height IS NOT NULL;
Solution:
SELECT * FROM (
SELECT
t.id,
ts.species,
t.height,
AVG(t.height) OVER (PARTITION BY ts.species) as avg_species_height,
STDDEV(t.height) OVER (PARTITION BY ts.species) as stddev_species_height,
COUNT(*) OVER (PARTITION BY ts.species) as species_count,
(t.height - AVG(t.height) OVER (PARTITION BY ts.species)) /
NULLIF(STDDEV(t.height) OVER (PARTITION BY ts.species), 0) as z_score
FROM trees t
JOIN taxonomy tx ON t.taxonomy_id = tx.id
JOIN tree_species ts ON tx.species_id = ts.id
WHERE t.height IS NOT NULL
) outlier_analysis
WHERE ABS(z_score) > 2
AND stddev_species_height IS NOT NULL
AND species_count >= 30
ORDER BY ABS(z_score) DESC;
Exercise 7: Distribution Analysis - Percentile Ranks for Tree Diameter
Goal: Calculate what percentage of trees in each domain have a smaller diameter than each tree.
Concepts: PERCENT_RANK(), CUME_DIST(), understanding percentiles
Step-by-step guide:
- Join trees with tree_domains
- Use
PERCENT_RANK()to get relative rank (0 to 1) - Use
CUME_DIST()to get cumulative distribution - Convert to percentages for easier interpretation
- Filter for valid diameter and domain values
Hints:
PERCENT_RANK()returns 0 for the smallest value, 1 for the largestCUME_DIST()returns the fraction of rows <= current row- Multiply by 100 to convert to percentages
- These help you understand distribution within groups
Solution:
SELECT
t.id,
td.domain,
t.diameter,
PERCENT_RANK() OVER (PARTITION BY td.domain ORDER BY t.diameter) as pct_rank,
ROUND(100 * PERCENT_RANK() OVER (PARTITION BY td.domain ORDER BY t.diameter), 2) as percentile,
CUME_DIST() OVER (PARTITION BY td.domain ORDER BY t.diameter) as cum_dist,
ROUND(100 * CUME_DIST() OVER (PARTITION BY td.domain ORDER BY t.diameter), 2) as pct_at_or_below
FROM trees t
JOIN tree_domains td ON t.domain_id = td.id
WHERE t.diameter IS NOT NULL
AND td.domain IS NOT NULL
ORDER BY td.domain, t.diameter DESC
LIMIT 200;
Exercise 8: Multi-Level Ranking - Best Trees by Species within Each Arrondissement
Goal: Create a two-level ranking showing the tallest trees of each species within each arrondissement.
Concepts: Multiple window functions, complex partitioning
Step-by-step guide:
- Join trees, locations, taxonomy, and tree_species
- Create a ranking by height within each (arrondissement, species) combination
- Also show how this tree ranks across all species in that arrondissement
- Filter to show only top 3 per species per arrondissement
- Use subquery to filter on window function results
Hints:
- You can have multiple
OVER()clauses with differentPARTITION BYconditions - First partition:
PARTITION BY arrondissement, species - Second partition:
PARTITION BY arrondissement - This shows both local (within species) and global (within area) rankings
Query Skeleton:
SELECT * FROM (
SELECT
...,
ROW_NUMBER() OVER (PARTITION BY arr, species ORDER BY height DESC) as rank_in_species,
ROW_NUMBER() OVER (PARTITION BY arr ORDER BY height DESC) as rank_in_area
FROM ...
JOIN ... ON ...
WHERE ...
) ranked
WHERE rank_in_species <= 3;
Solution:
SELECT * FROM (
SELECT
t.id,
l.arrondissement,
ts.species,
t.height,
ROW_NUMBER() OVER (PARTITION BY l.arrondissement, ts.species
ORDER BY t.height DESC) as rank_within_species,
ROW_NUMBER() OVER (PARTITION BY l.arrondissement
ORDER BY t.height DESC) as rank_in_arrondissement,
COUNT(*) OVER (PARTITION BY l.arrondissement, ts.species) as species_count_in_area
FROM trees t
JOIN locations l ON t.location_id = l.id
JOIN taxonomy tx ON t.taxonomy_id = tx.id
JOIN tree_species ts ON tx.species_id = ts.id
WHERE t.height IS NOT NULL
AND l.arrondissement IS NOT NULL
AND ts.species IS NOT NULL
) ranked_trees
WHERE rank_within_species <= 3
AND species_count_in_area >= 10
ORDER BY arrondissement, species, rank_within_species;
Exercise 9: Data Quality - Identifying Imbalanced Remarkable Trees
Goal: Analyze the distribution of the remarkable flag, showing how imbalanced it is across different domains and stages.
Concepts: Conditional aggregation with window functions, handling boolean/NULL values
Step-by-step guide:
- Join trees with tree_domains and tree_stages
- Count total trees per domain/stage combination
- Count trees where
remarkable = true - Count trees where
remarkable IS NULL - Calculate percentages for each category
- Use
DISTINCTto avoid row repetition
Hints:
COUNT(CASE WHEN remarkable = true THEN 1 END)counts only TRUE valuesCOUNT(CASE WHEN remarkable IS NULL THEN 1 END)counts NULLs- Window functions with CASE statements are powerful for conditional counting
- This reveals data quality issues with the remarkable flag
Query Skeleton:
SELECT DISTINCT
domain,
stage,
COUNT(*) OVER (PARTITION BY ...) as total,
COUNT(CASE WHEN ... THEN 1 END) OVER (PARTITION BY ...) as remarkable_count,
COUNT(CASE WHEN ... IS NULL THEN 1 END) OVER (PARTITION BY ...) as null_count,
-- percentages
FROM ...
ORDER BY ...;
Solution:
SELECT DISTINCT
td.domain,
ts.stage,
COUNT(*) OVER (PARTITION BY td.domain, ts.stage) as total_trees,
COUNT(CASE WHEN t.remarkable = true THEN 1 END) OVER (PARTITION BY td.domain, ts.stage) as remarkable_count,
COUNT(CASE WHEN t.remarkable IS NULL THEN 1 END) OVER (PARTITION BY td.domain, ts.stage) as null_count,
ROUND(100.0 * COUNT(CASE WHEN t.remarkable = true THEN 1 END) OVER (PARTITION BY td.domain, ts.stage) /
COUNT(*) OVER (PARTITION BY td.domain, ts.stage), 2) as pct_remarkable,
ROUND(100.0 * COUNT(CASE WHEN t.remarkable IS NULL THEN 1 END) OVER (PARTITION BY td.domain, ts.stage) /
COUNT(*) OVER (PARTITION BY td.domain, ts.stage), 2) as pct_null
FROM trees t
LEFT JOIN tree_domains td ON t.domain_id = td.id
LEFT JOIN tree_stages ts ON t.stage_id = ts.id
ORDER BY total_trees DESC;
Exercise 10: Running Totals - Cumulative Tree Count by Circumference Range
Goal: Create a running total showing how many trees exist at or below each circumference threshold within each genus.
Concepts: SUM() with window frames, running totals, ROWS BETWEEN
Step-by-step guide:
- Join trees, taxonomy, and tree_genres
- Order trees by circumference within each genus
- Use
SUM(1) OVER (PARTITION BY genus ORDER BY circumference ROWS BETWEEN UNBOUNDED PRECEDING AND CURRENT ROW) - This creates a running count
- Round circumferences to groups (e.g., multiples of 10) for clearer analysis
Hints:
- Window frames control which rows are included in the calculation
ROWS BETWEEN UNBOUNDED PRECEDING AND CURRENT ROWmeans “from start to here”- This is useful for cumulative distributions
SUM(1)is the same asCOUNT(*)
Query Skeleton:
SELECT
...,
SUM(1) OVER (PARTITION BY ...
ORDER BY ...
ROWS BETWEEN UNBOUNDED PRECEDING AND CURRENT ROW) as running_total
FROM ...
WHERE ...
ORDER BY ...;
Solution:
SELECT
t.id,
tg.genre,
t.circumference,
ROUND(t.circumference / 10.0) * 10 as circumference_bucket,
SUM(1) OVER (PARTITION BY tg.genre
ORDER BY t.circumference
ROWS BETWEEN UNBOUNDED PRECEDING AND CURRENT ROW) as running_total,
COUNT(*) OVER (PARTITION BY tg.genre) as total_in_genus,
ROUND(100.0 * SUM(1) OVER (PARTITION BY tg.genre
ORDER BY t.circumference
ROWS BETWEEN UNBOUNDED PRECEDING AND CURRENT ROW) /
COUNT(*) OVER (PARTITION BY tg.genre), 2) as pct_at_or_below
FROM trees t
JOIN taxonomy tx ON t.taxonomy_id = tx.id
JOIN tree_genres tg ON tx.genre_id = tg.id
WHERE t.circumference IS NOT NULL
AND tg.genre IS NOT NULL
ORDER BY tg.genre, t.circumference
LIMIT 500;
Exercise 11: Advanced - LAG and LEAD for Comparing Adjacent Trees
Goal: For trees ordered by height within each species, compare each tree to the previous and next tree.
Concepts: LAG(), LEAD(), accessing adjacent rows
Step-by-step guide:
- Join trees, taxonomy, and tree_species
- Use
LAG(height) OVER (PARTITION BY species ORDER BY height)to get previous tree’s height - Use
LEAD(height) OVER (PARTITION BY species ORDER BY height)to get next tree’s height - Calculate differences to see gaps in the distribution
- Filter to show interesting cases (large gaps)
Hints:
LAG(column, offset)looks backoffsetrows (default is 1)LEAD(column, offset)looks forwardoffsetrows- These return NULL at boundaries (first/last rows)
- Useful for identifying unusual gaps in sequential data
Solution:
SELECT * FROM (
SELECT
t.id,
ts.species,
t.height,
LAG(t.height) OVER (PARTITION BY ts.species ORDER BY t.height) as prev_height,
LEAD(t.height) OVER (PARTITION BY ts.species ORDER BY t.height) as next_height,
t.height - LAG(t.height) OVER (PARTITION BY ts.species ORDER BY t.height) as gap_from_prev,
LEAD(t.height) OVER (PARTITION BY ts.species ORDER BY t.height) - t.height as gap_to_next,
COUNT(*) OVER (PARTITION BY ts.species) as total_species_trees
FROM trees t
JOIN taxonomy tx ON t.taxonomy_id = tx.id
JOIN tree_species ts ON tx.species_id = ts.id
WHERE t.height IS NOT NULL
AND ts.species IS NOT NULL
) gaps
WHERE total_species_trees >= 50
AND (gap_from_prev > 5 OR gap_to_next > 5)
ORDER BY species, height;
Exercise 12: Complex Analysis - Identifying Stage Distribution Anomalies
Goal: Analyze the confusing stage labels by showing how stage distribution varies across domains and highlighting unusual patterns.
Concepts: Multiple window functions, conditional logic, complex filtering
Step-by-step guide:
- Join trees with tree_stages and tree_domains
- Calculate total trees per domain
- Calculate trees per stage within each domain
- Calculate percentage distribution
- Show which stages appear in which domains
- Use window functions to identify domains with unusual stage distributions
Hints:
- Stage values are inconsistent: “adult”, “young”, “young & adult”, and NULLs
- This exercise helps identify data quality issues
- Multiple PARTITION BY clauses can show different perspectives
- DISTINCT helps when showing summary statistics
Query Skeleton:
SELECT DISTINCT
domain,
stage,
COUNT(*) OVER (PARTITION BY domain, stage) as count_domain_stage,
COUNT(*) OVER (PARTITION BY domain) as count_domain,
COUNT(*) OVER (PARTITION BY stage) as count_stage,
-- percentages and other metrics
FROM ...
ORDER BY ...;
Solution:
SELECT DISTINCT
td.domain,
COALESCE(ts.stage, 'NULL/Missing') as stage,
COUNT(*) OVER (PARTITION BY td.domain, ts.stage) as trees_in_domain_stage,
COUNT(*) OVER (PARTITION BY td.domain) as total_in_domain,
COUNT(*) OVER (PARTITION BY ts.stage) as total_in_stage,
ROUND(100.0 * COUNT(*) OVER (PARTITION BY td.domain, ts.stage) /
COUNT(*) OVER (PARTITION BY td.domain), 2) as pct_of_domain,
ROUND(100.0 * COUNT(*) OVER (PARTITION BY td.domain, ts.stage) /
COUNT(*) OVER (), 2) as pct_of_all_trees,
COUNT(DISTINCT ts.stage) OVER (PARTITION BY td.domain) as distinct_stages_in_domain
FROM trees t
LEFT JOIN tree_domains td ON t.domain_id = td.id
LEFT JOIN tree_stages ts ON t.stage_id = ts.id
WHERE td.domain IS NOT NULL
ORDER BY td.domain, trees_in_domain_stage DESC;
Exercise 13: Master Challenge - Top Varieties with Complete Statistics
Goal: For each arrondissement, find the top 3 most common tree varieties and provide comprehensive statistics including rankings, percentages, and comparisons.
Concepts: Everything combined - multiple window functions, rankings, aggregations, complex joins
Step-by-step guide:
- Join all necessary tables: trees, locations, taxonomy, tree_varieties
- Count trees per variety per arrondissement
- Rank varieties within each arrondissement by count
- Calculate statistics: avg height, max circumference, etc.
- Calculate what percentage of the arrondissement’s trees each variety represents
- Filter to top 3 varieties per arrondissement
- Use subquery structure to handle complex filtering
Hints:
- This combines multiple concepts from previous exercises
- Use
DENSE_RANK()to handle ties appropriately - Multiple window functions with different PARTITION BY clauses
- Think about what statistics would be most useful for each variety
Query Skeleton:
SELECT * FROM (
SELECT
arrondissement,
variety,
COUNT(*) OVER (PARTITION BY arrondissement, variety) as variety_count,
DENSE_RANK() OVER (PARTITION BY arrondissement ORDER BY COUNT(*) OVER (PARTITION BY arrondissement, variety) DESC) as variety_rank,
AVG(...) OVER (PARTITION BY arrondissement, variety) as avg_stat,
COUNT(*) OVER (PARTITION BY arrondissement) as total_in_arrond,
-- more statistics
FROM trees t
JOIN ... ON ...
WHERE ...
) ranked_varieties
WHERE variety_rank <= 3
ORDER BY ...;
Solution:
SELECT * FROM (
SELECT DISTINCT
l.arrondissement,
tv.variety,
COUNT(*) OVER (PARTITION BY l.arrondissement, tv.variety) as variety_count,
DENSE_RANK() OVER (PARTITION BY l.arrondissement
ORDER BY COUNT(*) OVER (PARTITION BY l.arrondissement, tv.variety) DESC) as variety_rank,
COUNT(*) OVER (PARTITION BY l.arrondissement) as total_trees_in_arrond,
ROUND(100.0 * COUNT(*) OVER (PARTITION BY l.arrondissement, tv.variety) /
COUNT(*) OVER (PARTITION BY l.arrondissement), 2) as pct_of_arrondissement,
ROUND(AVG(t.height) OVER (PARTITION BY l.arrondissement, tv.variety), 1) as avg_height,
MAX(t.circumference) OVER (PARTITION BY l.arrondissement, tv.variety) as max_circumference,
COUNT(CASE WHEN t.remarkable = true THEN 1 END) OVER (PARTITION BY l.arrondissement, tv.variety) as remarkable_count
FROM trees t
JOIN locations l ON t.location_id = l.id
JOIN taxonomy tx ON t.taxonomy_id = tx.id
JOIN tree_varieties tv ON tx.variety_id = tv.id
WHERE l.arrondissement IS NOT NULL
AND tv.variety IS NOT NULL
) ranked_varieties
WHERE variety_rank <= 3
ORDER BY arrondissement, variety_rank;
Bonus Exercise 14: NTILE - Dividing Trees into Height Quartiles
Goal: Divide trees within each genus into 4 equal groups (quartiles) based on height to understand distribution.
Concepts: NTILE(), quantile analysis
Step-by-step guide:
- Join trees, taxonomy, and tree_genres
- Use
NTILE(4)to divide trees into 4 groups - Show which quartile each tree belongs to
- Calculate statistics for each quartile
Hints:
NTILE(n)divides rows into n approximately equal groups- Returns values 1, 2, 3, 4 for quartiles
- Useful for analyzing distribution across ranges
- Can use with different values: NTILE(10) for deciles, NTILE(100) for percentiles
Solution:
SELECT DISTINCT
tg.genre,
quartile,
MIN(height) OVER (PARTITION BY tg.genre, quartile) as min_height_in_quartile,
MAX(height) OVER (PARTITION BY tg.genre, quartile) as max_height_in_quartile,
ROUND(AVG(height) OVER (PARTITION BY tg.genre, quartile), 2) as avg_height_in_quartile,
COUNT(*) OVER (PARTITION BY tg.genre, quartile) as trees_in_quartile
FROM (
SELECT
t.*,
tg.genre,
t.height,
NTILE(4) OVER (PARTITION BY tg.genre ORDER BY t.height) as quartile
FROM trees t
JOIN taxonomy tx ON t.taxonomy_id = tx.id
JOIN tree_genres tg ON tx.genre_id = tg.id
WHERE t.height IS NOT NULL
AND tg.genre IS NOT NULL
) quartile_data
ORDER BY genre, quartile;
Summary of Window Functions Covered
- Aggregate functions:
COUNT(),SUM(),AVG(),MAX(),MIN(),STDDEV() - Ranking functions:
ROW_NUMBER(),RANK(),DENSE_RANK(),NTILE() - Distribution functions:
PERCENT_RANK(),CUME_DIST() - Offset functions:
LAG(),LEAD() - Conditional aggregation:
COUNT(CASE WHEN ... END) - Window frames:
ROWS BETWEEN UNBOUNDED PRECEDING AND CURRENT ROW
Each exercise builds on the previous ones, introducing new concepts while reinforcing earlier ones. Practice these queries, experiment with different window function combinations, and try creating your own variations!